inDISCO: INterpretable DIStributed COllaborative learning for biomedical images
Deep learning is fascinating . You can find its algorithms in the heart of self-driving cars, digital assistants, apps that make a photo look like an intergalactic selfie and in many other technological advances of the recent years. In healthcare, deep learning helps medical practitioners and scientists to analyse routine records, make a diagnosis from medical images, study biosignal and genomic data, and facilitate drug discovery [1].
Strict privacy policy often keeps medical data from a classical centralized learning (i.e. when all the data are on one device). A promising solution is DIStributed COllaborative (DISCO) learning which allows several data owners (clients) to learn a joint model without sharing data.
Privacy in DISCO learning, however, comes at cost to data transperancy resulting in clients learning blindly from their collegues. Now imagine if some of these collegues have hidden systemic bias in their dataset. We call this situation low data interoperablity and it often happens in the real-world. In this case, a final jointly trained model may not generalize well across all the participants. Another common issue with the state-of-the-art deep neural networks is their poor interpretability, i.e. we don't know why a model predicts what it predicts.
But what if we can build an algorithm to explore the data used in DISCO learning in an interpretable user-friendly way? What if we go further and claim that we can do it without compromizing clients' private data? Developing such an algorithm was the goal of my Master Thesis at the iGH group at EPFL and is a topic of my current research. Let's have a look at what we suggest.
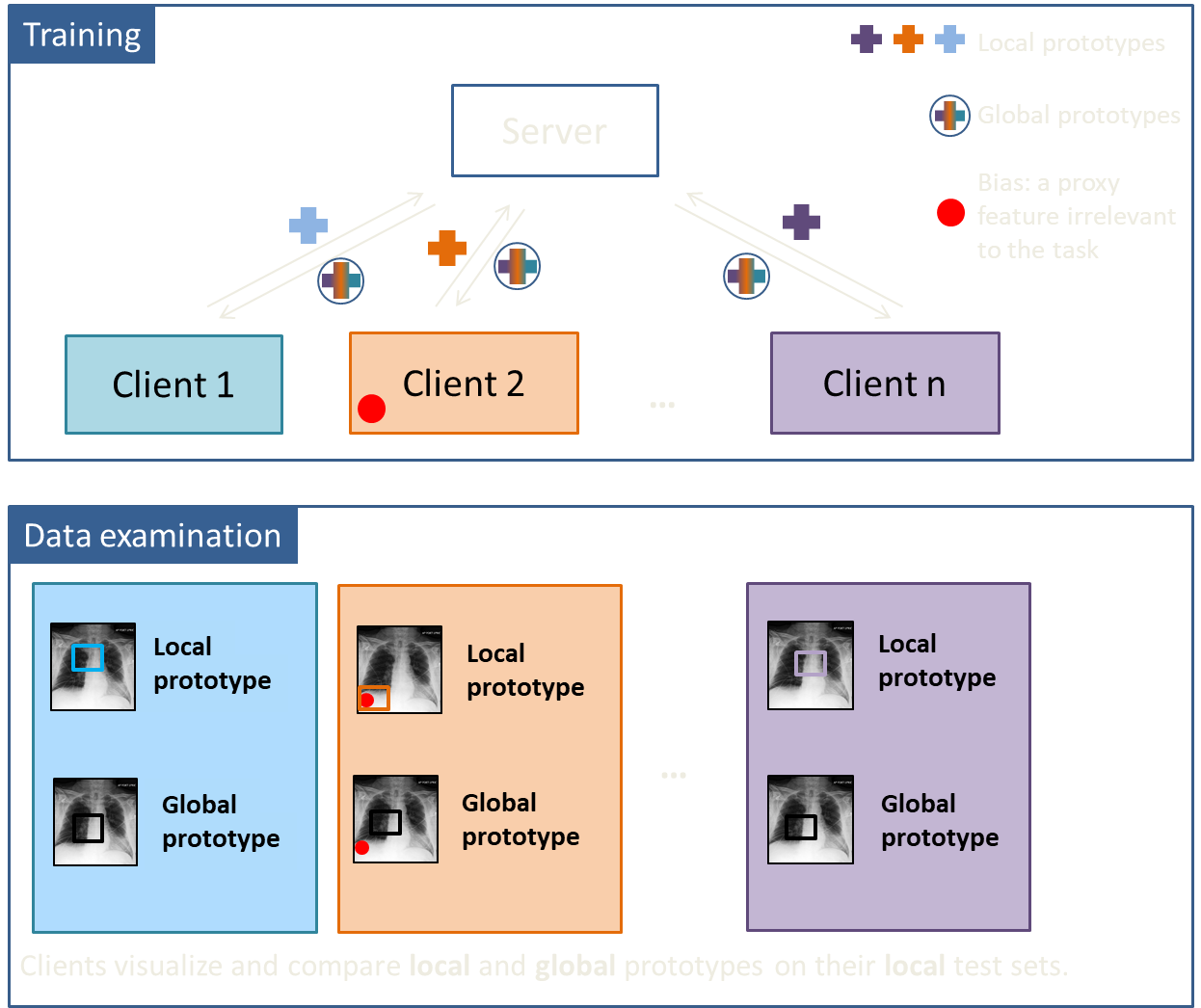
We were inspired from the work of Chen et al [2] and adapted their prototyical part learning network (ProtoPNet) to a DISCO setting. ProtoPNet is an interpretable-by-design network that has a human-friendly reasoning component. It bases its prediction on a similarity score between a test image and a set of learned prototypes for each category. With the help of this network, we developed an algorithm to identify a biased client in a privacy-preserving way. Within our inDISCO (INterpretable DIStributed COllaborative) approach, clients learn local prototypes on their private datasets and global ones via communication with each other.
After training, each client can compare local and global prototypes on its own test set by means of prototype visualization, i.e. by finding regions in the images most similar to the learned prototypes. In case of bias, there will be large diference between the global and local prototypes.
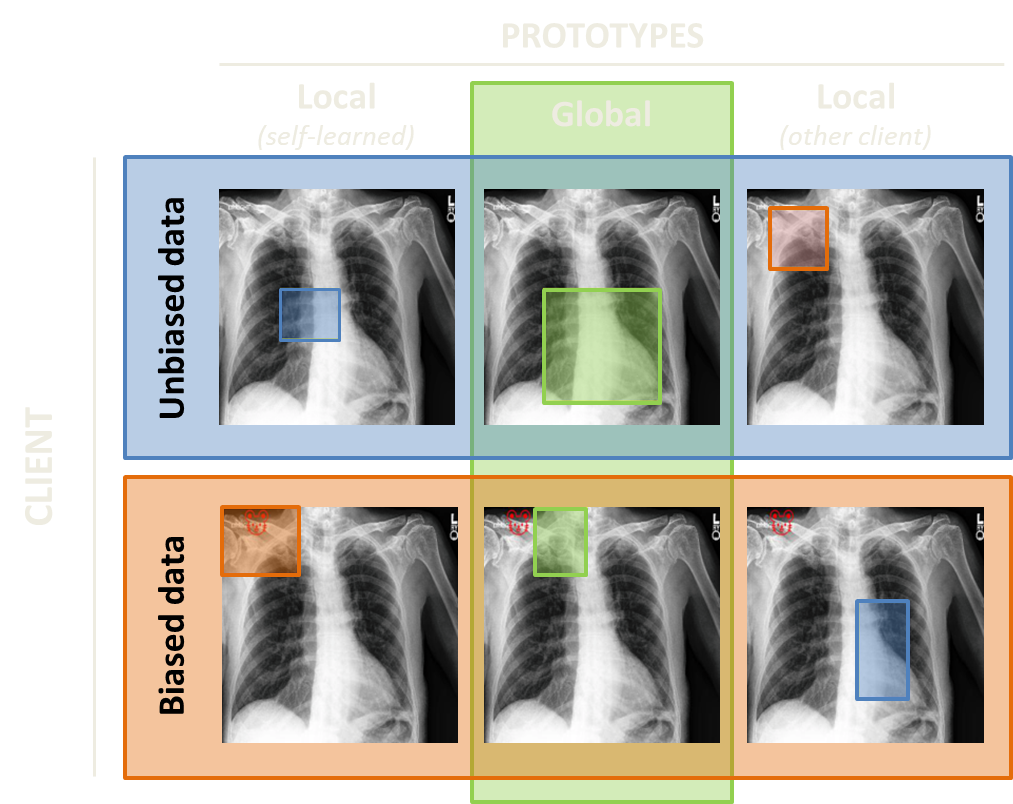
We demostrated our approach on classifying cardiomegaly and pleural effusion in the human X-ray images [3]. Cardiomegaly is a health condition characterized by an enlarged heart. Just as us, humans, would do, both local and global models learn this heart or its parts as prototypes for cardiomegaly class in the absence of data bias (first row, first two columns). To experiment with a simple bias, we added a small red emoji of a mouse in the left upper corner of the images in the cardiomegaly class of one of the clients. This little mouse has changed the game! Now, a local model trained solely on biased data learns an amoji as a prototype (second row, first column). However, a global model which learns its prototypes in communication with unbiased clients doesn't look at the emoji and prefers the neighboring region. This visual difference between local and global prototypes indicates the presence of incompatible data in the federation.
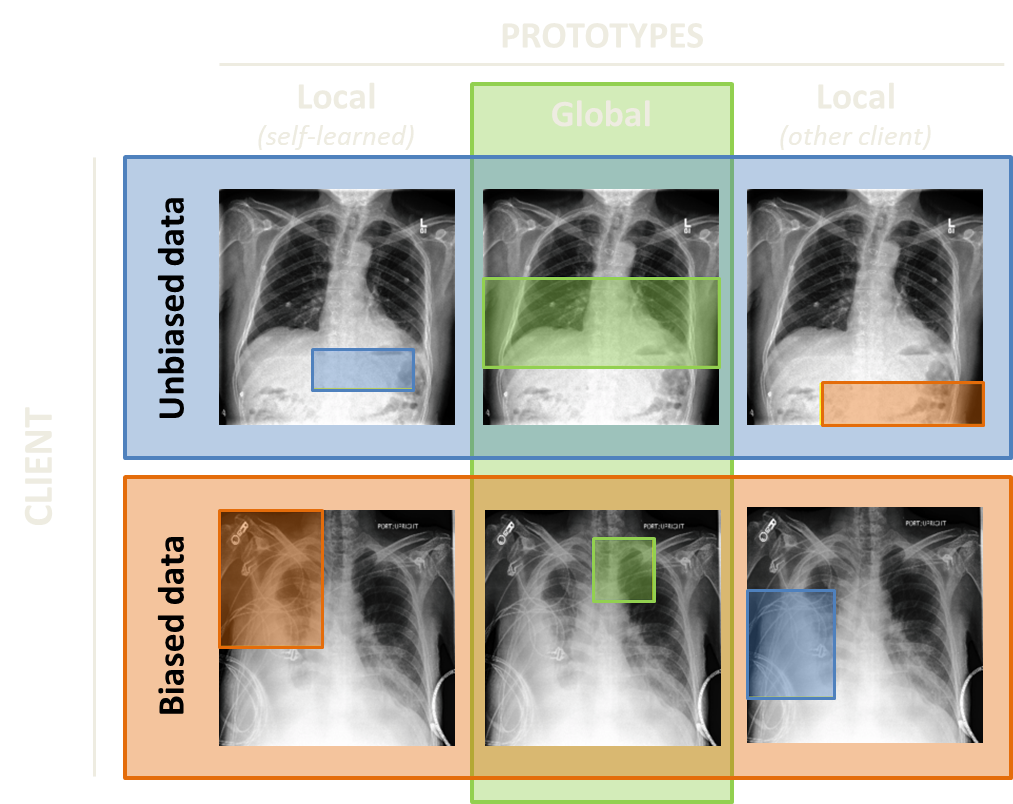
Pleural effusion is an accumulation of fluid between the visceral and parietal pleural membranes that line the lungs and chest cavity. For this class, unbiased prototypes are mostly bottom part of the lungs. Since patients with pleural effusion often get chest drains to eliminate the fluid from their lungs, these drains can bias the prediction of pleural effusion. You can see this situation on the right: prototype in the first column second row is far from the bottom of the lungs because a local model relies on the presence of chest drain to predict pleural effusion and not on the presence of fluid in the lungs. And again, there is a clear difference between local and global prototypes in a biased case (second row, second column).
Our current work is to study the trade-off between privacy and ease of bias identification by varying the amount of network parameters to share. This will allow clients to decide how much information they are ready to communicate depending on their data sensitivity.
To be continued
Supervisers: Prof. Mary-Anne Hartley, Prof. Martin Jaggi
External advisor: Dr. Sai Praneeth Karimireddy